Monarch® Spin RNA Cleanup Kit Protocol
This protocol is for the Monarch Spin RNA Cleanup Kits (NEB #’s NEB #T2030, T2040, and T2050). The protocol can also be used with the columns for RNA Cleanup (NEB #’s T2037, T2047, and T2057).
The standard protocol outlined below will purify RNA ≥ 25 nt. A simple modification in Step 2 can allow for the purification of RNA as small as 15 nt.
Before You Begin:
- Add 4 volumes of ethanol (≥ 95%) to the Monarch Monarch Buffer WX before use, as directed on the bottle.
- All centrifugation steps should be carried out at 16,000 x g. (~13K RPM in a typical microcentrifuge). This ensures all traces of buffer are eluted at each step.
Protocol Steps:
- Add 100 μl Monarch Buffer BX to the 50 μl sample. A starting sample volume of 50 μl is recommended. For smaller samples, nuclease-free water can be used to adjust the volume. For samples larger than 50 μl, scale buffer volumes accordingly. Samples with a starting volume > 150 μl will require reloading of the column during Step 3.
- Add 150 μl (1 volume) of ethanol (≥ 95%) to your sample and mix by pipetting or flicking the tube. Do not vortex. This will enable the binding of RNA ≥ 25 nt. If you wish to bind RNA as small as 15 nt, add 2 volumes (300 μl) of ethanol to your sample instead of 1 volume (150 μl). The addition of 2 volumes of ethanol shifts the cutoff size of RNA binding from 25 nt down to 15 nt.
- Insert column into collection tube, load sample onto column and close the cap. Spin for 1 minute, then discard flow-through. For diluted samples > 900 μl, load a portion of the sample, spin, and then repeat as necessary.
 To save time, spin for 30 seconds, instead of 1 minute.
To save time, spin for 30 seconds, instead of 1 minute.
- Re-insert column into collection tube. Add 500 μl Monarch Buffer WX and spin for 1 minute. Discard the flow-through.
 To save time, spin for 30 seconds, instead of 1 minute.
To save time, spin for 30 seconds, instead of 1 minute.
- Repeat wash (Step 4).
- Transfer column to an RNase-free 1.5 ml microfuge tube (not provided). Use care to ensure that the tip of the column does not come into contact with the flow-through. If in doubt, re-spin for 1 minute to ensure traces of salt and ethanol are not carried over to next step.
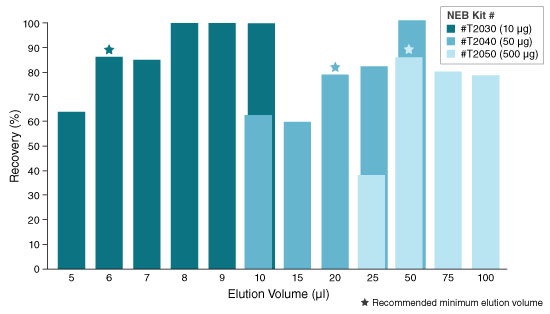
- Elute in nuclease-free water according to the table below. The eluted RNA can be used immediately or stored at -70°C. Care should be used to ensure the elution buffer is delivered onto the matrix and not the wall of the column to maximize elution efficiency.
KIT ELUTION VOLUME INCUBATION TIME SPIN TIME T2030 6–20 µl N/A 1 minute T2040 20–100 µl N/A 1 minute T2050** 50–100 µl 5 minutes (Room temp.) 1 minute
* When cleaning up large amounts of RNA (> 100 μg, NEB #T2050), some precipitation may occur following the addition of the Monarch Buffer BX and ethanol to the sample (Steps 1 and 2). A pellet containing the RNA of interest may form on the side of the column following the first binding spin (Step 3). To maximize recovery of this RNA, a second elution is recommended.
** Yield may slightly increase if a larger volume is used, but the RNA will be less concentrated.
 To save time, spin for 30 seconds, instead of 1 minute.
To save time, spin for 30 seconds, instead of 1 minute.
Additional Resources:
- Product Manual
- Troubleshooting Guide for RNA Cleanup
- Visit NEBMonarch.com for information on these and other Monarch Kits

