NEBNext® Magnesium RNA Fragmentation Module Protocol
Protocol
Starting Material
Total RNA (2–50 μg) or purified mRNA (50–250 ng)
- Mix the following components in a sterile PCR tube:
COMPONENT VOLUME (µl) Purified RNA 1–18 RNA Fragmentation Buffer (10X) 2 Nuclease-Free Water variable Total volume 20 - Incubate in a preheated thermal cycler for 1–5 minutes at 94°C.*
- Transfer tube to ice.
- Add 2 μl 10X RNA Fragmentation Stop Solution.
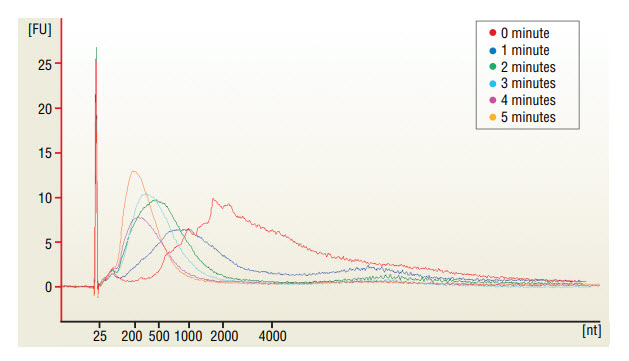
* Fragmentation time should be adjusted depending on amount and type of RNA and desired sizes of fragments (See Figure 1).
Clean Up Fragmented RNA Using the Monarch RNA Cleanup Kit (NEB #T2030)
- Add 78 μl of the Nuclease-Free Water to the 22 μl fragmented RNA from step 4. Purify sample using the Monarch® RNA Cleanup Kit (NEB #T2030) following manufacturer instructions. Elute in 14.5 µl Nuclease-Free Water. The recovered volume should be ~13.5 µl.
Note: column purification removes short RNA Fragments and enriches the sample for RNA fragments longer than 200 nucleotides.
Alternatively, Clean Up Fragmented RNA Using Ethanol Precipitation
- Mix the following components in a sterile 1.5 ml microcentrifuge tube:
COMPONENT VOLUME (µl) Fragmented RNA from Step 4 22 3M Sodium Acetate, pH 5.2 2 Linear Acrylamide, 10 mg/ml 1–2 100% Ethanol 60 Total volume 85–86 - Incubate at -80°C for 30 minutes.
- Centrifuge at 14,000 rpm for 25 minutes at 4°C in a microcentrifuge.
- Carefully remove ethanol.
- Wash pellet with 300 μl of 70% ethanol.
- Centrifuge and carefully remove 70% ethanol.
- Air dry pellet for up to 10 minutes at room temperature to remove residual ethanol.
- Resuspend in 13.5 μl Nuclease-Free Water.
Assess the Yield and the Size Distribution of the Fragmented RNA.
Take 1 μl of the fragmented RNA and dilute it 1:10 with nuclease-free water. Run 1 μl in the Agilent Bioanalyzer 2100 using a RNA Pico chip (Figure 1).