RNA Synthesis Protocols (E2070)
Standard RNA Synthesis
1. Thaw necessary kit components, mix and pulse spin in a microfuge to collect solutions at the bottom of the tubes. Keep on ice.
2. If you are planning to set up many reactions, it is convenient to prepare a master mix by combining equal volumes of the SP6 Reaction Buffer (10X) and each of the four ribonucleotide (NTP) solutions. Use 12.5 µl of master mix per reaction.
3. Assemble the reaction at room temperature in the following order:
| Nuclease-free water |
X µl |
|
| SP6 Reaction Buffer (10X) |
2.5 µl |
|
| ATP (Tris), 50 mM |
2.5 µl |
5 mM final |
| GTP (Tris), 50 mM |
2.5 µl |
5 mM final |
| UTP (Tris), 50 mM |
2.5 µl |
5 mM final |
| CTP (Tris), 50 mM |
2.5 µl |
5 mM final |
| Template DNA |
X µl |
1 µg |
| SP6 RNA Polymerase Mix |
2.5 µl |
|
| Total reaction volume |
25 µl |
4. Mix thoroughly and pulse-spin in a microfuge. Incubate at 37°C for 2 hours.
For reaction times of 60 minutes or less, a water bath or heat block may be used. For reaction times longer than 60 minutes we recommend using a dry air incubator or thermocycler to prevent evaporation/condensation.
5. Optional: DNase treatment to remove DNA template. Standard reactions normally generate large amounts of RNA at high concentrations. As a result, the reaction mixture may be quite viscous. It is easier to perform DNase treatment after the reaction mixture is diluted with water. To remove template DNA, add 25 µl nuclease-free water to each 25 µl reaction followed by 2 µl of DNase I (RNase-free). Mix and incubate for 15 minutes at 37°C.
6. Proceed with purification of synthesized RNA or analysis of transcription products by gel electrophoresis.
Capped RNA Synthesis
The recommended ratio of cap analog to GTP is 4:1. Cap analogs are sold separately. Please refer to the ordering information section or www.neb.com for more information.
1. Thaw necessary kit components, mix and pulse-spin in a microfuge to collect solutions at the bottom of the tubes. Keep on ice.
2. Make a 10 mM GTP solution by diluting an aliquot of 50 mM GTP 1:5 with nuclease-free water.
3. Prepare the cap analog at 40 mM concentration.
4. Assemble the reaction at room temperature in the following order:
| Nuclease-free water |
X µl |
|
| SP6 Reaction Buffer (10X) |
2.5 µl |
|
| ATP (Tris), 50 mM |
2.5 µl |
5 mM final |
| UTP (Tris), 50 mM |
2.5 µl |
5 mM final |
| CTP (Tris), 50 mM |
2.5 µl |
5 mM final |
| GTP (Tris), 10 mM |
2.5 µl |
1 mM final |
| Cap Analog (40 mM) |
2.5 µl |
4 mM final |
| Template DNA |
X µl |
1 µg |
| SP6 RNA Polymerase Mix |
2.5 µl |
|
| Total reaction volume |
25 µl |
5. Mix thoroughly, pulse-spin in microfuge and incubate at 37°C for 30 minutes.
Table 1. Effect of cap analog:GTP ratios on RNA yield.
| CAP ANALOG: GTP RATIO | CONCENTRATION OF CAP ANALOG:GTP (mM) | RNA YIELD (µG) | % CAPPED RNA |
|---|---|---|---|
| 0:1 | 0:5 | 100 | 0 |
| 1:1 | 2.5:2.5 | 70 | 50 |
| 2:1 | 3.3:1.7 | 50 | 67 |
| 4:1 | 4:1 | 30 | 80 |
| 8:1 | 4.4:0.56 | 5–15 | 89 |
6. Optional: To remove the DNA template, add 25 µl of water and 2 µl of DNase I (RNase-free), mix and incubate at 37°C for 15 minutes.
7. Proceed with purification of synthesized RNA or analysis of transcription products by gel electrophoresis.
RNA Synthesis with Modified Nucleotides
Dye-labeled, Biotin-labeled or other modified NTPs are not supplied with this kit. The recommended molar ratio of modified NTP [Biotin-, Fluorescein-, Digoxigenin-, Aminoallyl-NTPs (or others)] to standard NTP is 1:3 or 1:2. The following reaction set up assumes that modified UTP is used.
1. Thaw necessary kit components, mix and pulse-spin in microfuge to collect solutions at the bottom of the tubes. Keep on ice.
2. Assemble the reaction at room temperature in the following order:
| Nuclease-free Water |
X µl |
|
| SP6 Reaction Buffer (10X) |
2.5 µl |
|
| ATP (Tris), 50 mM |
2.5 µl |
5 mM final |
| GTP (Tris), 50 mM |
2.5 µl |
5 mM final |
| CTP (Tris), 50 mM |
2.5 µl |
5 mM final |
| UTP (Tris), 50 mM |
1.75 µl |
3.5 mM final |
| Modified UTP (10 mM) |
3.75 µl |
1.5 mM final |
| Template DNA |
X µl |
1 µg |
| SP6 RNA Polymerase Mix |
2.5 µl |
|
| Total reaction volume |
25 µl |
Note that the ratio of UTP:modified UTP can be adjusted to meet specific needs. The total amount of UTP can be lowered if higher RNA yield is not necessary.
3. Mix thoroughly, pulse-spin in a microfuge and incubate at 37°C for 2 hours. For short transcripts, incubate at 37°C for 4–16 hours.
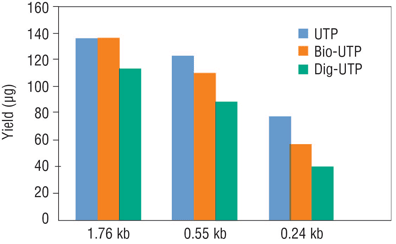
Modified ribonucleotides reduce transcription efficiency, therefore lower yields should be expected as compared to transcription using unmodified NTPs (Figure 4). In general, Biotin-NTP and Aminoallyl-NTP have less of an effect on yields, while lower yields can be expected for transcription reactions containing Fluorescein-NTP, Cy-NTP or Digoxigenin-NTP. In addition, transcripts containing modified ribonucleotides have reduced electrophoretic mobility due to higher molecular weight.
4. Optional: To remove the DNA template, add 25 µl of water and 2 µl of DNase I (RNase-free), mix and incubate at 37°C for 15 minutes.
5. Proceed with purification of synthesized RNA or analysis of transcription products by gel electrophoresis.
High Specific Activity Radiolabeled RNA Probe Synthesis
The HiScribe SP6 RNA Synthesis Kit can be used to synthesize high specific activity radiolabeled RNA probes by following the modified protocol below. More than 50% of the label can be incorporated in a 10 minute reaction. The labeled RNA probes have a specific activity of approximately 108 cpm/µg.
1. Choosing 32P labeled nucleotide:
We recommend using [α-32P] UTP or CTP at 800-6000 Ci/mmol and ≥ 10 mCi/ml for the synthesis of RNA labeled probes. We do not recommend using radiolabeled ATP or GTP since less label is generally incorporated. RNA labeled with [α-32P] ATP or GTP appears to be more subject to decomposition during storage.
2. Thaw the necessary kit components, mix and pulse-spin in microfuge to collect solutions to the bottom of the tubes. Keep on ice.
3. Dilute 50 mM UTP to 50 µM UTP in two steps as described below if [α-32P] UTP is used.
a. Prepare 200 µl 1 mM UTP by combining 4 µl 50 mM UTP and 196 µl nuclease-free water. Extra 1 mM UTP can be stored at –20°C for future use.
b. Prepare 100 µl 50 µM UTP by combining 5 µl 1 mM UTP and 95 µl nuclease-free water.
4. Prepare master mix. For accurate pipetting we recommend preparing 20 µl master mix, which is enough for 4 labeling reactions. The master mix contains reaction buffer, ATP, GTP and CTP.
| Nuclease-free Water |
9 µl |
| SP6 Reaction Buffer (10X) |
5 µl |
| ATP (Tris), 50 mM |
2 µl |
| GTP (Tris), 50 mM |
2 µl |
| CTP (Tris), 50 mM |
2 µl |
| Total master mix volume |
20 µl |
Use 4 µl master mix for each labeling reaction. Extra master mix can be stored at –20°C for future use.
5. Assemble the reaction at room temperature in the following order:
| Nuclease-free Water |
X µl |
|
| Master Mix (A, G, C, Buffer) |
4 µl |
0.8 mM each NTP, final |
| UTP (Tris) 50 µM |
3 µl |
6 µM final |
| (α-32P) UTP |
X µl |
0.13-2 µM final |
| Template DNA |
X µl |
0.1 to 1 µg |
| SP6 RNA Polymerase Mix |
2.5 µl |
|
| Total reaction volume |
25 µl |
The labeled NTP is present at a limiting concentration and is therefore referred to as the “limiting nucleotide”. The “limiting nucleotide” is a mixture of both the labeled and unlabeled form of that specific NTP. There is a trade-off between synthesis of high specific activity probe and synthesis of full-length probe. The higher the concentration of the “limiting nucleotide”, the higher the proportion of full-length transcripts, but if the unlabeled nucleotide is used to increase the “limiting nucleotide” concentration, it will lower the specific activity of the transcript. For most labeling reactions, 4-8 µM of the “limiting nucleotide" is necessary for full-length probe synthesis with high specific activity. The template sequence will also affect the specific activity. For example, if the transcript contains more UTP then more 32P-UTP will be incorporated resulting in a higher specific activity. For longer RNA transcripts > 1 kb it may be necessary to increase the concentration of the unlabeled “limiting nucleotide” to increase the proportion of full-length transcript, however the improvement in yield of full-length transcript will reduce the specific activity of the probe. We recommend increasing the concentration of the unlabeled “limiting nucleotide” to 20 µM.
Table 2. Concentration of [α-32P] NTP in a transcription reaction.
| SPECIFIC ACTIVITY (Ci/mmol) | CONCENTRATION (mCi/ml) | VOLUME USED PER 25 µl REACTION | CONCENTRATION IN 25 µl REACTION (HOT LABEL) |
|---|---|---|---|
| 800 | 10 | 1 µl |
0.5 µM |
| 800 | 20 | 1 µl |
1 µM |
| 800 | 40 | 1 µl | 2 µM |
| 3,000 | 10 | 1 µl |
0.13 µM |
| 3,000 | 20 | 1 µl | 0.27 µM |
| 3,000 | 40 | 1 µl |
0.53 µM |
| 6,000 | 40 | 1 µl | 0.27 µM |
6. Mix thoroughly, pulse-spin in a microfuge and incubate for 10 minutes. Incubation temperature is not crucial for labeling efficiency, room temperature to 40°C can be used.
7. Optional: To remove template DNA, add 25 µl of water and 2 µl of DNase I (RNase-free), mix and incubate for 15 minutes at 37°C.
8. Proceed with purification of synthesized RNA or analysis of transcription products by gel electrophoresis.