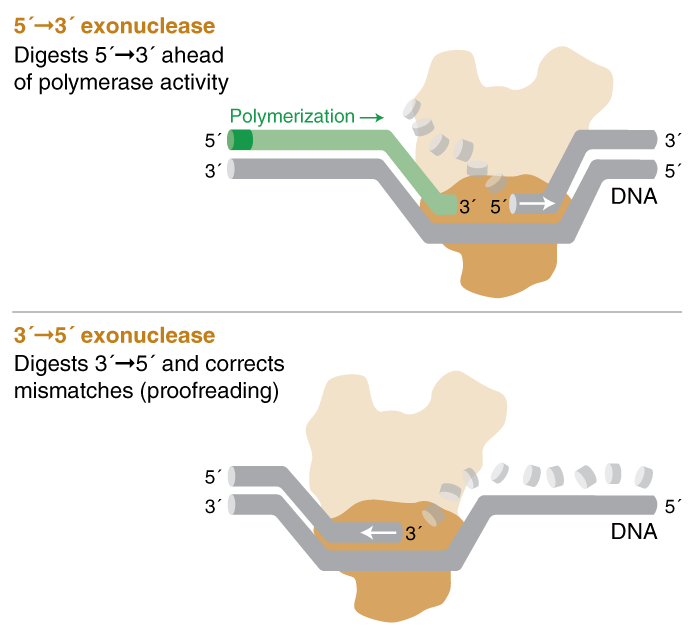
Polymerases can possess up to two types of exonuclease activity:
- 5´- 3´ exonuclease activity will digest nucleotides in the 5´ to 3´ direction on a template ahead of polymerase activity.
- Taq polymerase possesses a specific type of 5´-3´ exonuclease activity called 5' flap endonuclease activity, which catalyzes the cleavage of 5´ DNA flaps from a DNA duplex that has a single-stranded 5´ overhang on one of the strands.
- 3´ - 5´ exonuclease activity, AKA proofreading activity, digests nucleotides with 3´ hydroxyl groups from the 3´ to 5´ direction. This includes correction of mismatched base pairs and terminal 3´ digestion.
- Some polymerases possess one or both of these types of exonuclease activities.
- In the presence of dNTPs, polymerization activity is higher than exonuclease activity.
- Some polymerases possess neither of these activities.
Learn More
What does it mean when a polymerase has 5´- 3´ exonuclease activity?
When polymerases with 5´- 3´ exonuclease activity encounter a downstream nucleotide on the same strand, the phosphodiester bonds of the encountered strand are hydrolyzed (digested) ahead of polymerization.
Taq polymerase possesses a specific type of 5´- 3´ exonuclease activity called 5´ flap endonuclease activity, which catalyzes the cleavage of 5´ DNA flaps from a DNA duplex that has a single-stranded 5' overhang on one of the strands. When Taq polymerase encounters such a flap structure, such as the fluorophore on a qPCR hydrolysis probe, it sequentially digests the phosphodiester bonds between nucleotides in the 5´- 3´ direction ahead of polymerization.
The relationship between 3´ - 5´ exonuclease (proofreading) activity and PCR product length.
DNA polymerases ensure accurate replication using a series of molecular checkpoints at the site of nucleotide incorporation and beyond. During nucleotide addition, the correct incoming nucleotide is positioned for a productive alignment of catalytic groups, ensuring efficient incorporation. This alignment for catalysis is sensitive to distortions in position caused by incorrect Watson-Crick base pairing, allowing for kinetic stalling at incorrect or non-cognate base pairs.
Polymerases that possess proofreading activity “check” whether or not the correct nucleotide has been inserted into the template. If a mismatch is detected, the DNA is transferred from the polymerization domain to the N-terminal 3´ - 5´ exonuclease domain of the polymerase. The incorrectly incorporated nucleotide is excised, DNA moves back into the polymerization domain, and copying can resume.
The absence of 3´ - 5´ exonuclease (proofreading) activity may have ramifications other than fidelity in PCR. The lack of proofreading activity in Taq DNA Polymerase has been proposed to limit the maximum amplicon size possible. Generally, Taq performs best when amplifying DNA fragments < 2 kb but can work with fragments up to 3 – 4 kb. When kept to this amplicon size, Taq is a robust, easily optimized enzyme. However, with products above ~3 kb it quickly drops in effectiveness. During PCR, Taq will misincorporate nucleotides and produce mismatches, making it prone to stalling and more likely to dissociate before extending the full-length product.
The combination of a certain amplicon size and polymerase error rate can result in accumulation of enough mismatched 3´ ends to effectively inhibit the PCR process. These mismatched 3´ ends are particularly problematic for Taq because it lacks the 3´→ 5´ exonuclease activity to remove them. By mixing in a small amount of a polymerase with proofreading exonuclease activity, such as Deep Vent® DNA Polymerase, amplification of fragments ≥ 20 kb can be achieved (see LongAmp® DNA Polymerase). Since the vast majority of the enzyme in the blend is Taq DNA Polymerase, it is probably doing the bulk of the primer extension, with the proofreading Deep Vent DNA Polymerase removing the inhibitory 3´ mismatches generated by Taq.
To learn more about the anatomy of a polymerase and how structure relates to function, see this Feature Article.
How to block exonuclease activity during PCR? Add phosphorothioate linkages to primers.
A phosphorothioate (pt) bond is a phosphodiester linkage where one of the two non-bridging oxygens has been replaced by a sulfur atom, which inhibits nuclease phosphodiesterase activity. Chemically, the substitution of oxygen with sulfur does not dramatically change the reactivity of the bond, and pt-containing polynucleotides can still function in many enzymatic reactions. In most contexts, 2-3 pt bonds at the 3´ end are sufficient to block polymerase 3´-5´ exonuclease activity.
